Molecular dynamic simulations reveal detailed spike-ACE2 interactions
$ 12.99 · 5 (284) · In stock
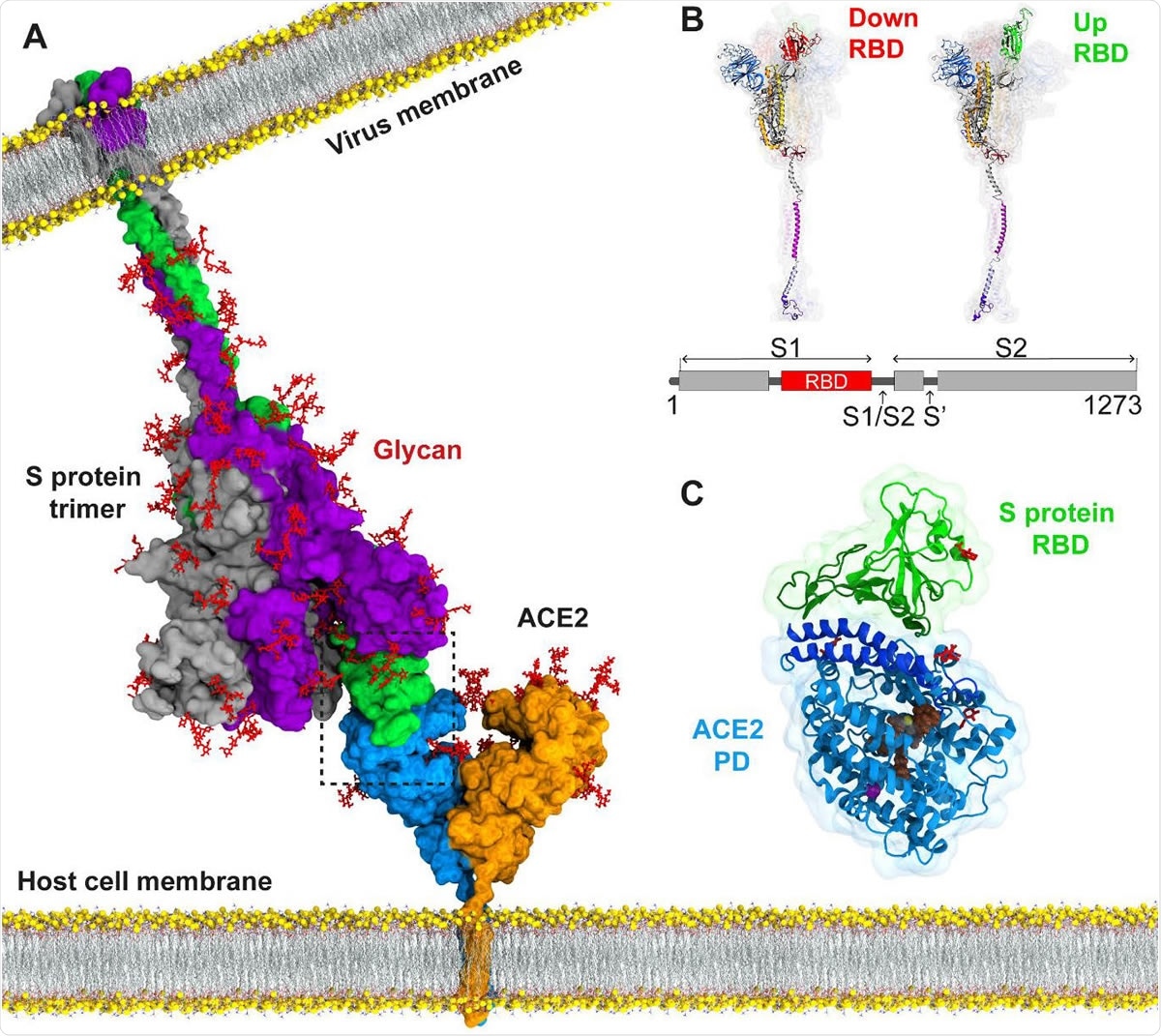
The current COVID-19 pandemic has spread throughout the world. Caused by a single-stranded RNA betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is closely related to but much more infectious than the earlier highly pathogenic betacoronaviruses SARS and MERS-CoV, has impacted social, economic, and physical health to an unimaginable extent.

Computational biophysical characterization of the SARS-CoV-2 spike protein binding with the ACE2 receptor and implications for infectivity - Computational and Structural Biotechnology Journal

Different compounds against Angiotensin-Converting Enzyme 2 (ACE2) receptor potentially containing the infectivity of SARS-CoV-2: an in silico study

Molecular dynamic simulation suggests stronger interaction of Omicron-spike with ACE2 than wild but weaker than Delta SARS-CoV-2 can be blocked by engineered S1-RBD fraction
Self-Association of ACE-2 with Different RBD Amounts: A Dynamic Simulation Perspective on SARS-CoV-2 Infection
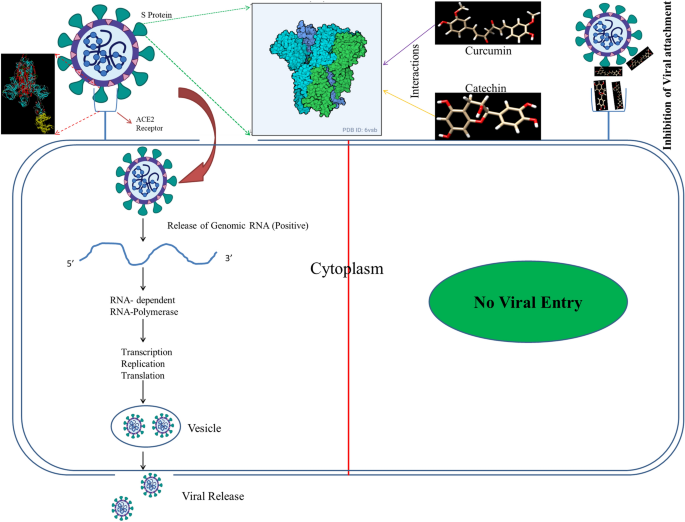
Catechin and curcumin interact with S protein of SARS-CoV2 and ACE2 of human cell membrane: insights from computational studies
An experimental peptide could block Covid-19, MIT News
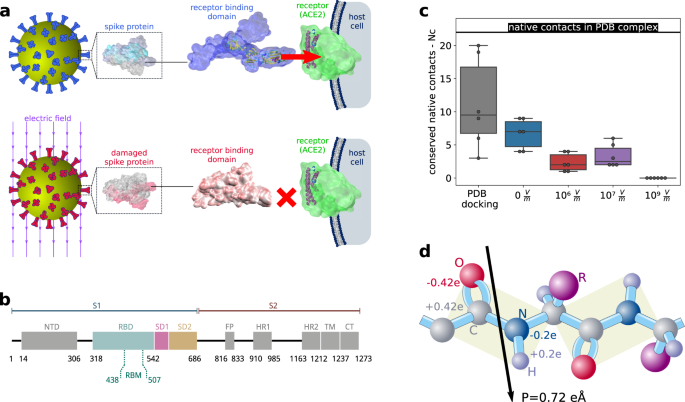
The SARS-CoV-2 spike protein is vulnerable to moderate electric fields

Biology, Free Full-Text

Molecular dynamics simulations highlight the altered binding landscape at the spike-ACE2 interface between the Delta and Omicron variants compared to the SARS-CoV-2 original strain - ScienceDirect
